Abstract
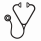
Introduction: Blinatumomab, a CD19/CD3 bi-specific T-cell engager monoclonal antibody that re-directs CD3-positive T cells towards CD19-positive B cells, has shown promise in the treatment of R/R B-cell precursor acute lymphoblastic leukemia (B-ALL), with superior survival rates compared to salvage chemotherapy. However, many patients do not respond or subsequently relapse, and the mechanisms underlying resistance are unclear. The goals of this study were to characterize the genomic features associated with response to blinatumomab.
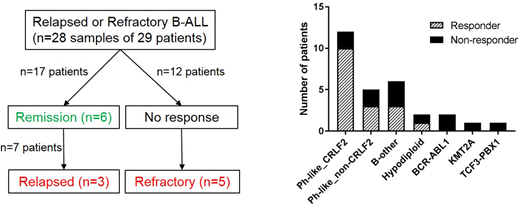
Methods: We studied 29 patients (pts; median age 28, range 18-70) with R/R B-ALL who were treated with up to 5 cycles of blinatumomab, and predominantly of Hispanic ancestry (66%). Overall, 17 pts (59%) achieved remission with blinatumomab whilst 12 showed no response. Among the 17 responders, 7 (41%) subsequently relapsed or progressed during treatment. We analyzed leukemic blasts obtained before and after blinatumomab treatment whenever available: pre-blinatumomab R/R (n=28), post-blinatumomab refractory (n=5), post-blinatumomab relapsed (n=3) (Figure 1). Leukemia and matched remission samples were studied using transcriptome sequencing (n=34), whole genome sequencing (n=28), whole exome sequencing (n=19) and Infinium Omni2.5Exome-8 (SNP array, n=19).
Results: Seventeen of 29 pts (59%) were Ph-like ALL. Twelve of 17 Ph-like ALL pts had high CRLF2 expression, among these we identified P2RY8-CRLF2 (n=4) and IGH-CRLF2 (n=8). Within the remaining 5 Ph-like ALL cases, two pts harbored NUP214-ABL1, two IGH-EPOR and one TERF-JAK2. Fifteen of the 17 (88%) Ph-like ALL cases were of Hispanic ancestry. The prevalence of other known subtypes was relatively low: BCR-ABL1 7%, hypodiploid 7%, KMT2A 3%, TCF3-PBX1 3% and B-other 21%. We observed a high response rate of 83.3% (10/12 cases) in Ph-like_CRLF2 pts, whilst the frequency of response was 60% (3/5, including two IGH-EPOR) for Ph-like_non-CRLF2 pts, and 33% (4/12) for the other subtypes (Ph-like ALL vs. others, P=0.029) (Figure 1). Unsupervised hierarchical clustering of pre-blinatumomab samples identified 3 clusters based on response to blinatumomab: cluster 1 contained non-responders, clusters 2 and 3 were largely made up of responders. By gene expression profiling using CIBERSORT we found reduced infiltration of cytotoxic CD8+ T-cells in cluster 1 compared to clusters 2 and 3 (6.1% vs. 14.9%, P=0.014), which was inversely correlated with the presence of CD4+ T cells (17.9% vs. 11.5%). GSEA showed enrichment for the IFNγ response, JAK-STAT signaling, chemokine and cytokine signaling in responders. In non-responders, differential gene expression analysis identified up-regulation of the H3K4 demethylase KDM5B, an oncogene associated with progression and chemoresistance of glioma and neuroblastoma. We observed a high frequency of alterations affecting B-lymphoid development (IKZF1, PAX5 and EBF1) in the pre-blinatumomab samples (20 of 22, 91%), which were maintained during progression or relapse. The frequency of B-lymphoid alterations did not differ significantly between responders and non-responders (13 of 14, 93% vs. 7 of 8, 88%). Alterations affecting the cell cycle (CDKN2A/B, TP53, RB1) were observed at a high frequency in pre-blinatumomab samples (15 of 22, 68%), with CDKN2A/B deletions enriched in responders compared to non-responders (11 of 14, 79% vs. 2 of 8, 25%; P=0.026). We also observed a high prevalence of alterations affecting epigenetic modifiers (ARID1B, CREBBP, KDM6A, KMT2D, TRRAP, SMARCA4) in pre-blinatumomab samples (17 of 22, 77%), with no difference between responders and non-responders (10 of 14, 71% vs. 7 of 8, 88%; P=0.61). Of the post-blinatumomab R/R samples available for study (n=8), CD19 expression was negative (n=1), dim (n=2) or positive (n=5). In contrast to previous reports of CD19 escape in CAR T-cell treated patients, there was no evidence of aberrantly spliced CD19 mRNA species, CD19 mutation or deletion in the three negative/dim cases.
Conclusion: We show that a heightened immune response through the infiltration of cytotoxic T-cells and activation of IFNγ and JAK-STAT signaling in leukemic cells is an important determinant of response to blinatumomab. Importantly, blinatumomab is a valid therapeutic approach for patients harboring high-risk CRLF2 and EPOR-rearrangements. CD19 escape is not associated with genetic alterations at the CD19 locus.
Stein:Amgen Inc.: Speakers Bureau; Celgene: Speakers Bureau. Mullighan:Loxo Oncology: Research Funding; Cancer Prevention and Research Institute of Texas: Consultancy; Amgen: Honoraria, Speakers Bureau; Pfizer: Honoraria, Research Funding, Speakers Bureau; Abbvie: Research Funding. Forman:Mustang Therapeutics: Other: Licensing Agreement, Patents & Royalties, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract